Darko Štern
Experience in image and signal analysis with a strong focus on machine learning. After a career shift, my research and professional interest is now focused on machine learning methods for testing and validation of ADAS/AD systems, as well as batteries and fuel-cells. Previos research interests are concentrated around the design and development of algorithms for processing and analysis of three-dimensional (3D) computed tomography (CT) and magnetic resonance (MR) images.
At AVL, we are constantly searching for motivated students interested in doing their master theses on the topic of ADAS/AD, battery, and fuel-cell testing. Please, visit our web page for more information or check the Project section bellow.
Experience
Tehnology Scout for Cognitive Testing
Senior Project Researcher
Senior Researcher
Marie Curie Research Fellow
Independent Researcher
Projects
I am constantly looking for students with a research interest in machine learning, image and signal processing in domains of ADAS/AD, and battery and fuel cell testing. This page lists specific open student projects at the master's or bachelor's level. Please, also check AVL web page.
Evaluation of AI/ML methods for cognitive testing of AD stack

Testing an AD stack in a virtual environment requires a cognitive testing methodology that will go beyond the full factorial variation of the parameters of all possible scenarios. The task of the student is to compare the performance and identify the limitations of Cognitive Testing methods developing at AVL with testing methods available in the literature on a publicly available AD stack (e.g. Autowave and Apollo).
List of all Projects !
Publications
List of my publication can also be found at Google Scholar and ReserchGate . If you have any problems accessing our publications, feel free to contact me.
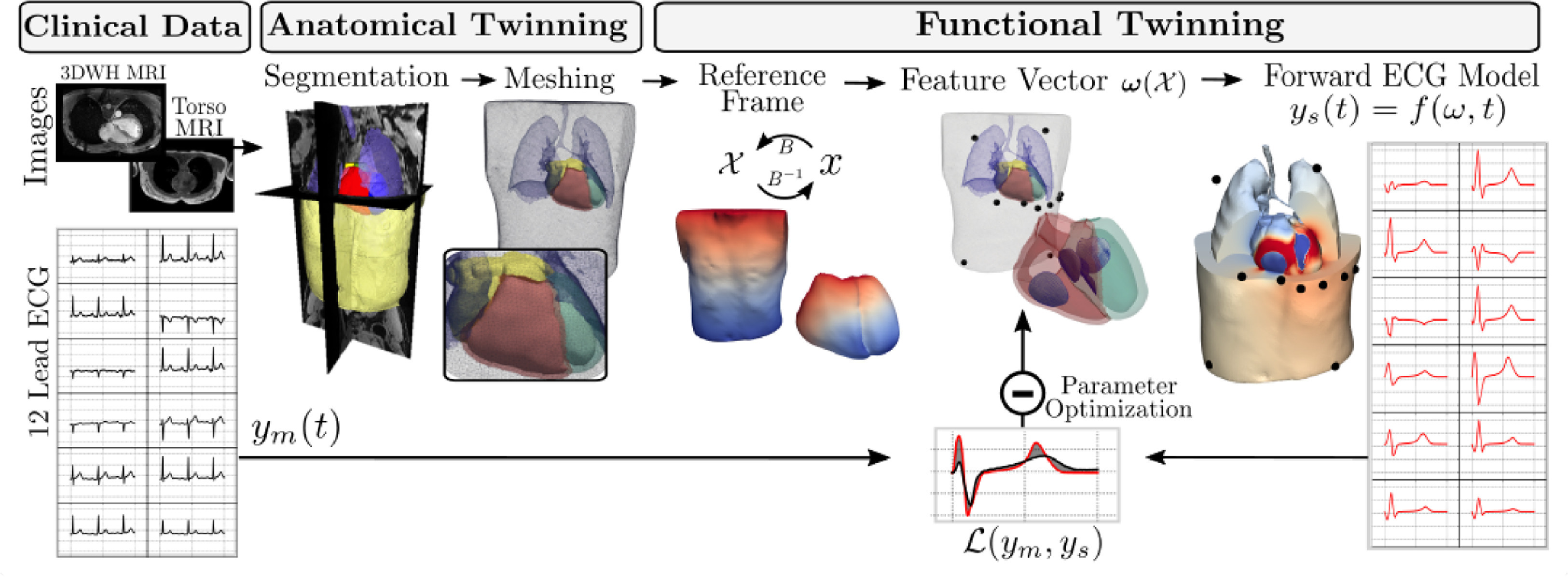
A Framework for the generation of digital twins of cardiac electrophysiology from clinical 12-leads ECGs
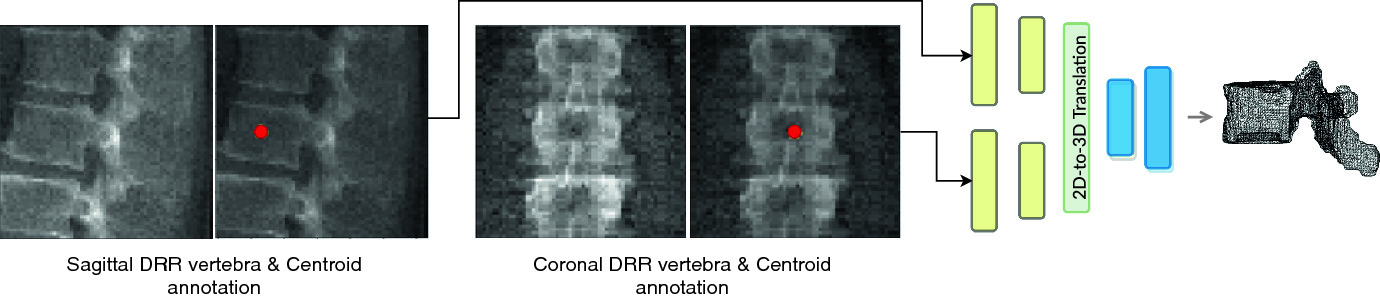
Inferring the 3D standing spine posture from 2D radiographs
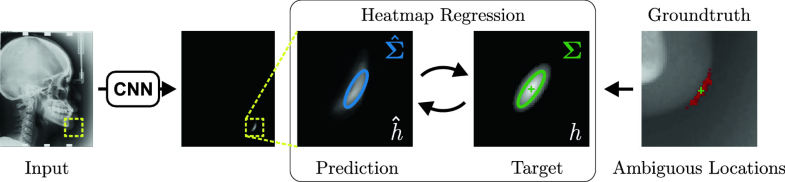
Uncertainty Estimation in Landmark Localization Based on Gaussian Heatmaps
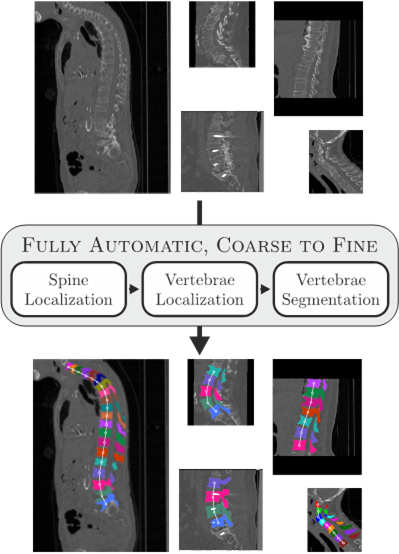
Coarse to Fine Vertebrae Localization and Segmentation with SpatialConfiguration-Net and U-Net
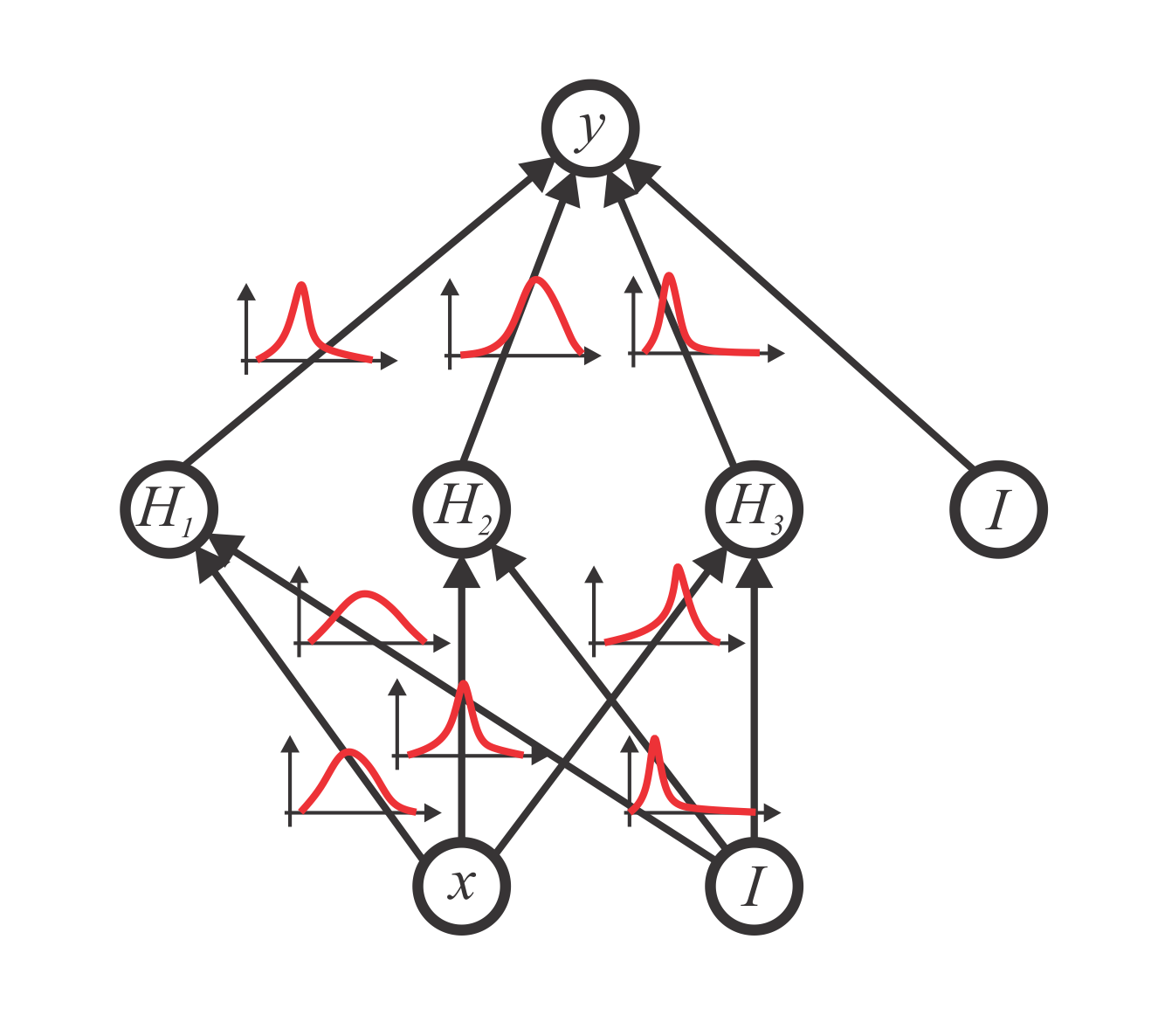
Variational Inference and Bayesian CNNs for Uncertainty Estimation in Multi-Factorial Bone Age Prediction
Evaluating Spatial Configuration Constrained CNNs for Localizing Facial and Body Pose Landmarks

Evaluation of algorithms for Multi-Modality Whole Heart Segmentation: An open-access grand challenge
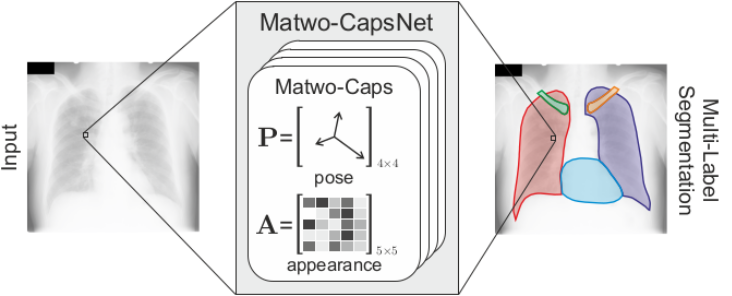
Matwo-CapsNet: A Multi-Label Semantic Segmentation Capsules Network
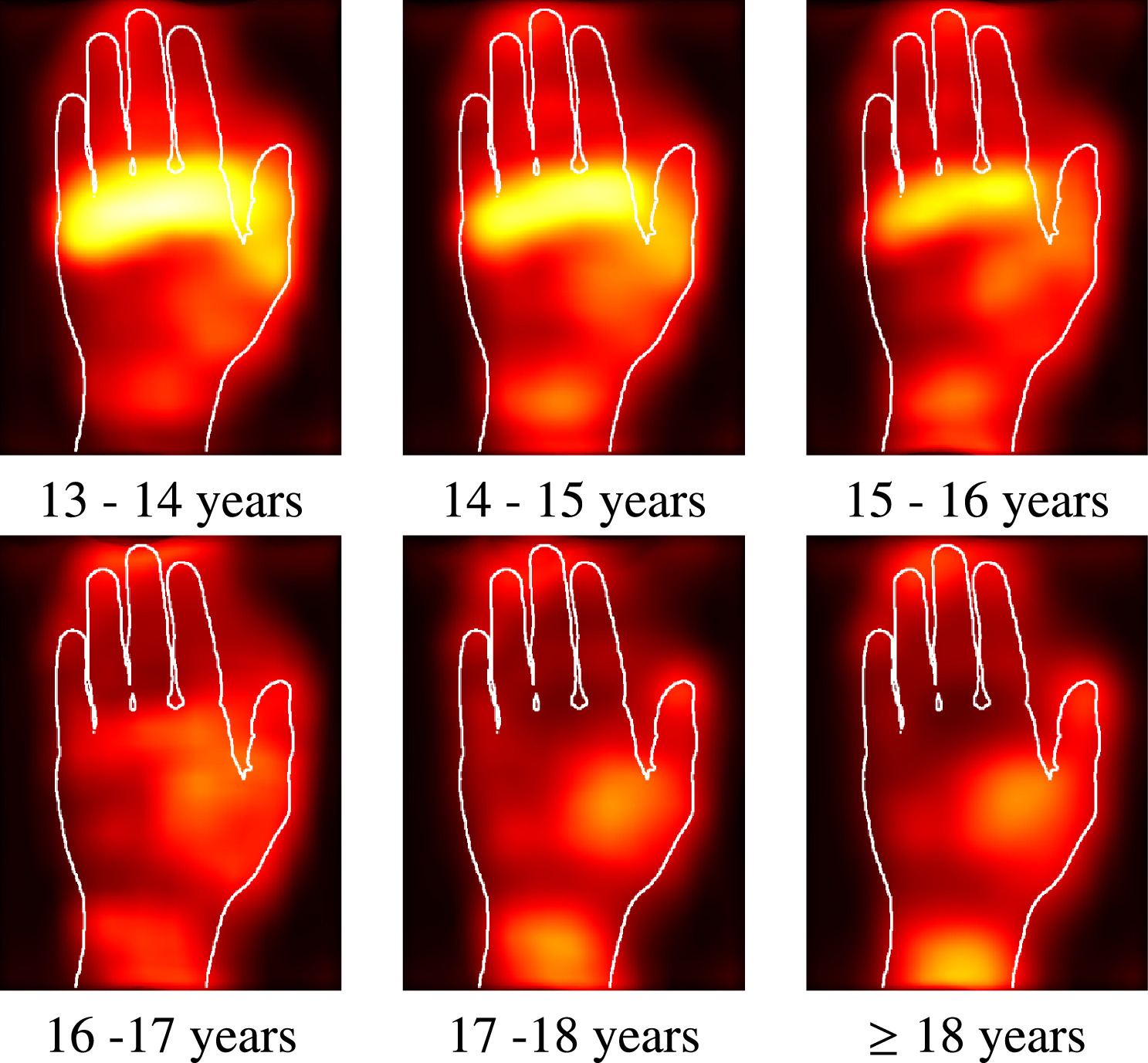
Automated age estimation from MRI volumes of the hand
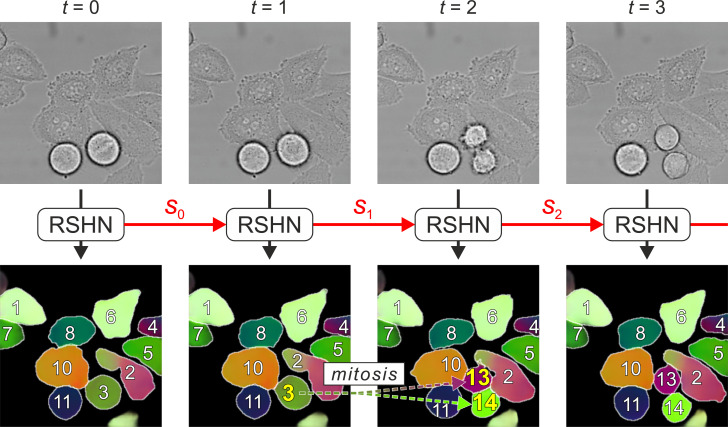
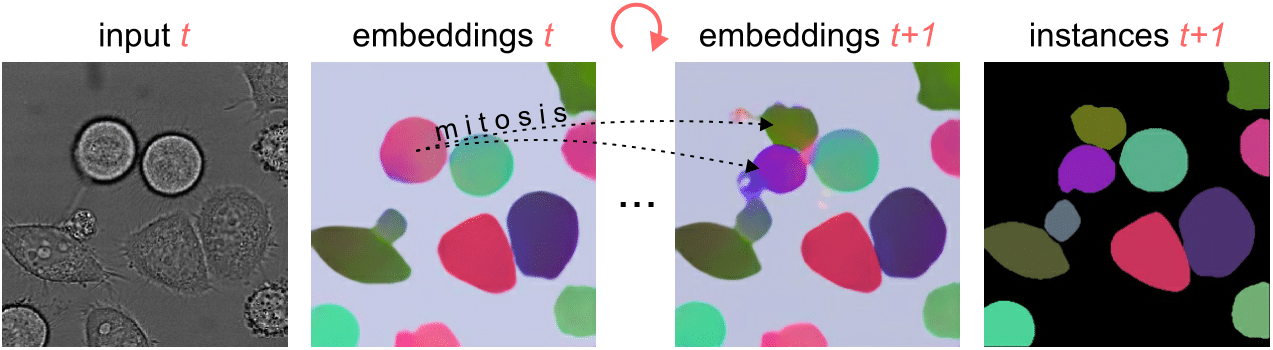
Segmenting and tracking cell instances with cosine embeddings and recurrent hourglass networks
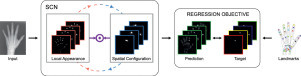
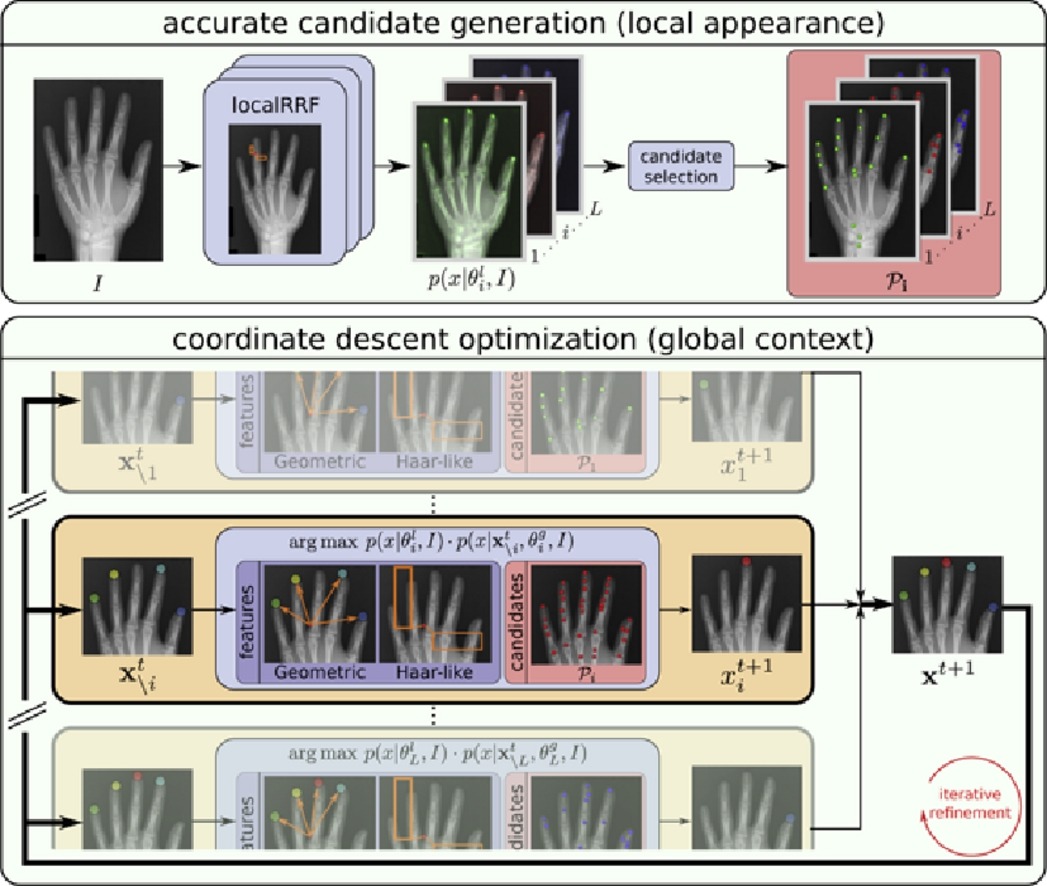
Integrating Spatial Configuration into Heatmap Regression Based CNNs for Landmark Localization
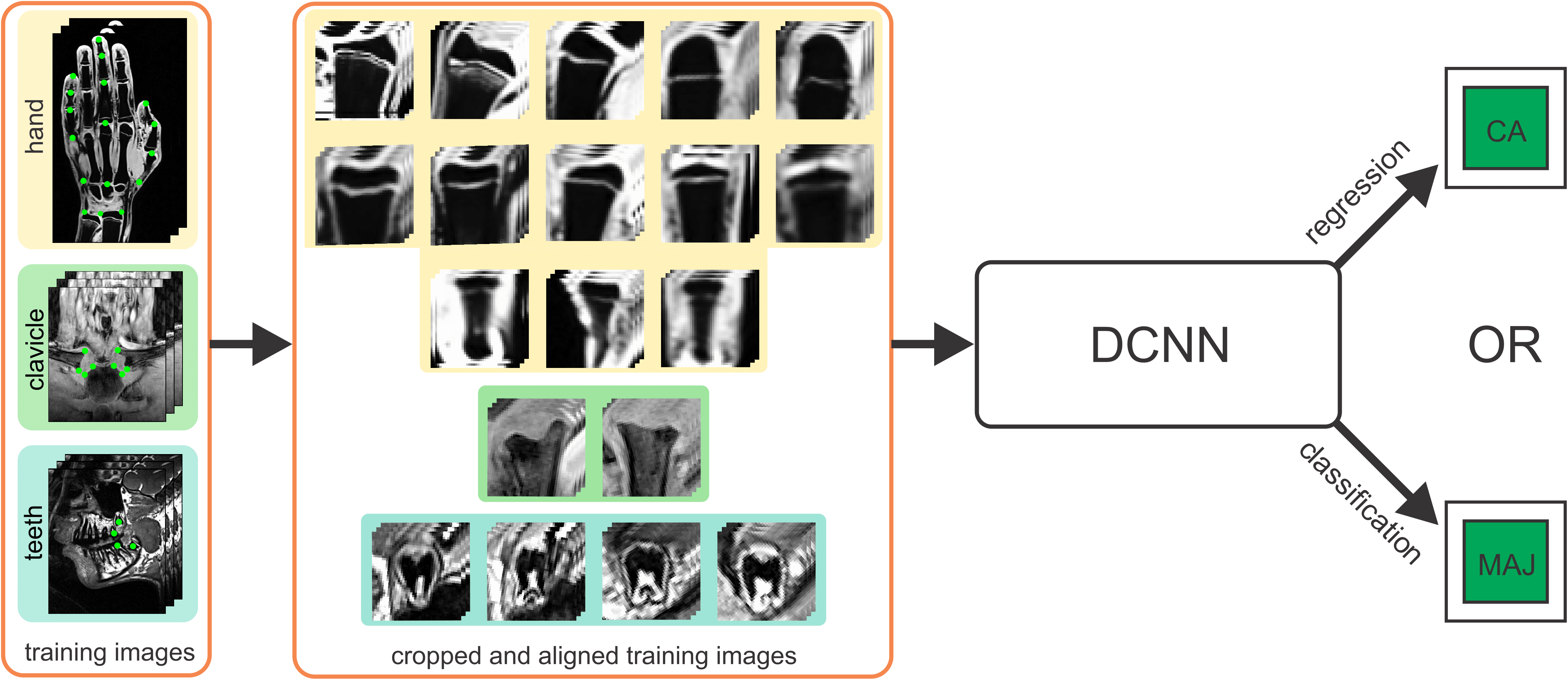
Automatic Age Estimation and Majority Age Classification from Multi-Factorial MRI Data
Sparse-View CT Reconstruction Using Wasserstein GANs
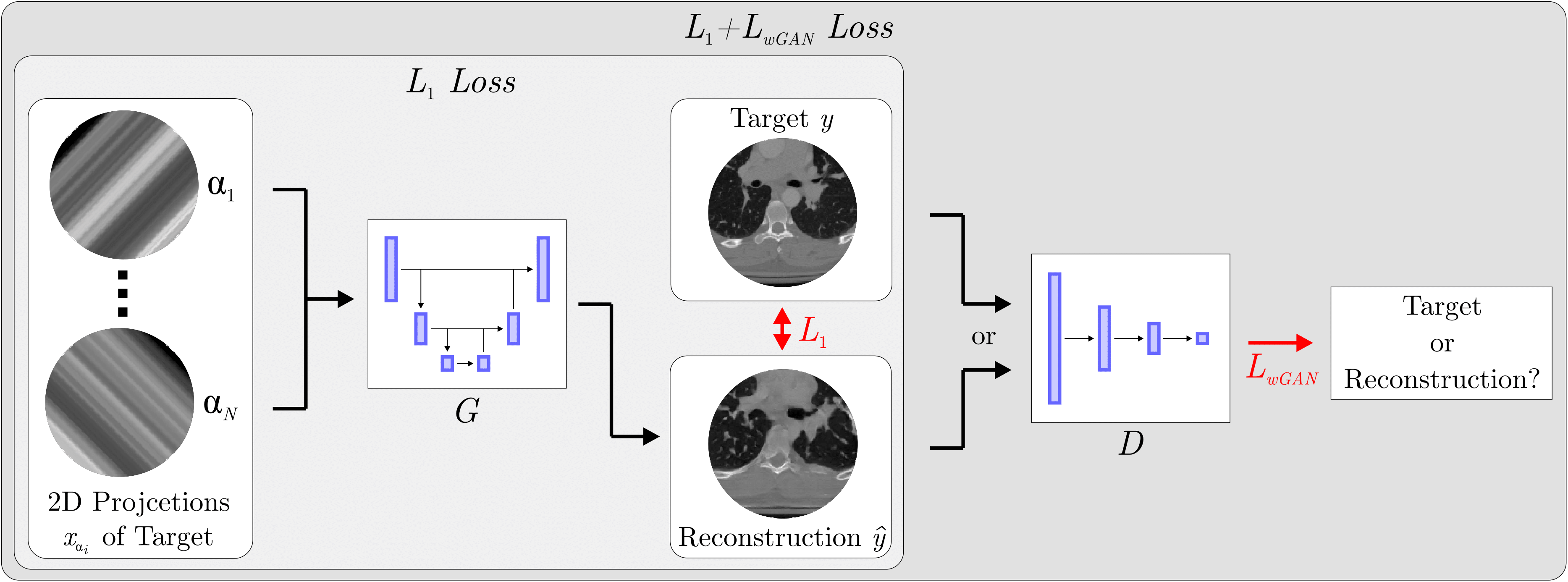
Instance segmentation and tracking with cosine embeddings and recurrent hourglass networks
Integrating geometric configuration and appearance information into a unified framework for anatomical landmark localization
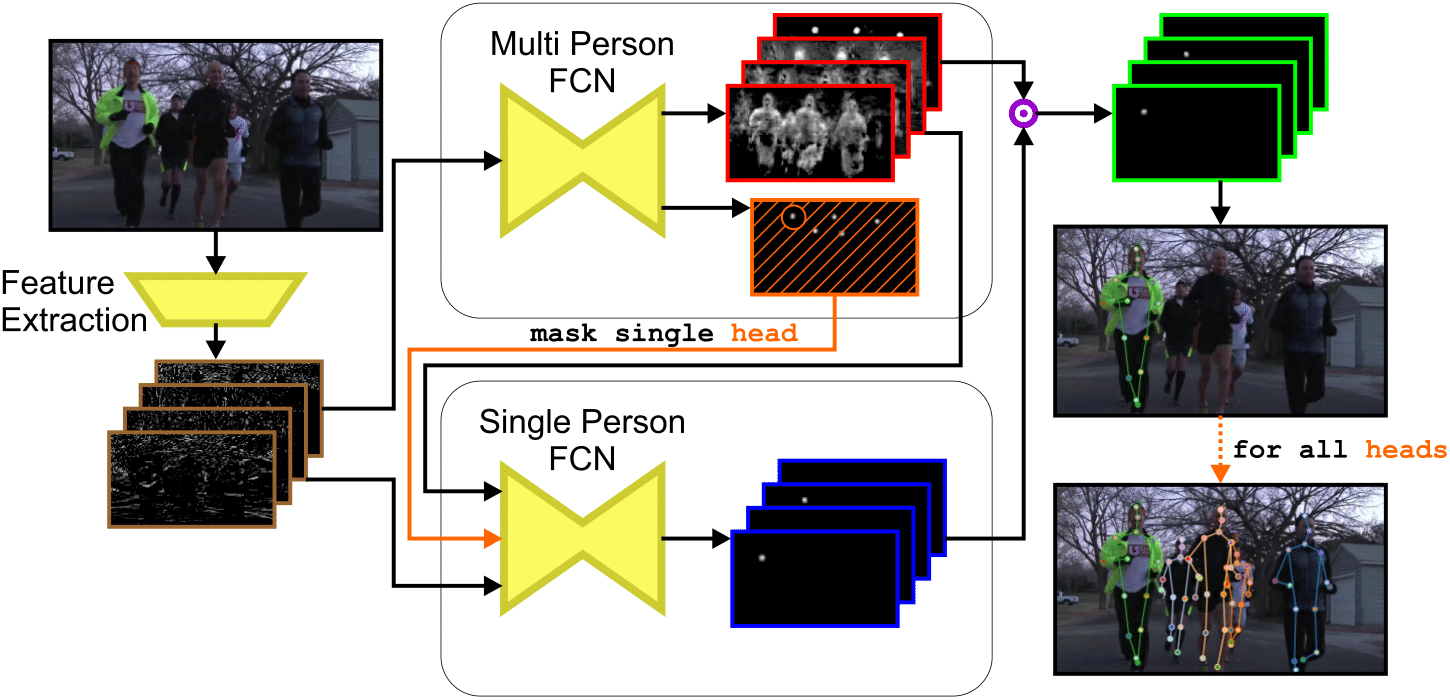
Simultaneous multi-person detection and single-person pose estimation with a single heatmap regression network